By Vince Giuliano
Methuselah –Methuselah lived 969 years according to Genesis 5:27, the oldest person who ever lived. Image source

I believe the previous blog entry YGB YOUNGING1.0 – THE EMERGING AGING REVERSAL STRATEGY may well turn out to be the most important one published in this blog so far. In that blog entry, Steve Buss and I described some key elements of a strategy for age reversal which appears to be emerging in the recent literature –we call it YOUNGING1.0. To our knowledge this is the most extensive scientific characterization of a viable age reversal strategy that has appeared in the literature.
If you are new to this YOUNGING series, I suggest you start with reviewing the background in the initial blog entry YGA Introduction to the YOUNGING Series – Emerging Aging Reversal Strategies and Treatments. Then go on to read YGB YOUNGING1.0 – THE EMERGING AGING REVERSAL STRATEGY before you tackle the material here. Finally, after reviewing this entry you might want to to on to reading YGD YOUNGING1.0 PART 4: UNDERLYING MECHANISMS OF YOUNGING 1.0. HSC STEM CELL DIFFERENTIATION.
This current blog entry covers clarification, expansion and evidence for some key points that were laid out in that original blog entry. In particular, I expand on the hypothesized scientific basis for YOUNGING1.0, which is the reversal of global H3K27 histone methylation via JMJD2/JMJD3. I go deeper into the biology and processes of histone methylation. And I selectively treat a few associated topics. For example, I note the role of vitamin C in both OSKM cell regression to younger states and inYOUNGING1.0 via JMJD2/3.
Again, this blog entry. I take a somewhat Socratic approach, identifying key questions that could come up and our responses to these questions – to the extent I am able to formulate them.
Steve bus and I have asserted that the key feature of the particular YOUNGING approach, we focused on, YOUNGING1.0 is demethylation of H3K27 via the demethylases JMJD2/3 Why? In the original blog entry we did not explicitly cover some of the basic reasons for this informed conjecture. The issue was brought to my attention via an email from a well-informed colleague in the longevity science field. He wrote me the following email.
I’ve been out of touch with you for awhile, and one of my readers just referred me to your recent work on Younging1.0. I’m glad to hear that you’re thriving and on the road to a younger body. I’d like to talk to you when you have a chance. My general response to the science you present is this: I think of epigenetics as a highly specific program, turning some genes on and others off, and involving many mechanisms, of which 5-methyl-cytosine and H3K27 methylation are just two examples. The problem as I see it is to selectively turn on the right genes and turn off the wrong ones. So I’m questioning whether JMJD2 can do anything selective, or whether its effect is to turn genes on globally. If you have time for a video chat, please let me know when it’s convenient.
We indeed did follow through with a video chat and I repeat here my responses to the points he raises here. I generally agree with what my colleague says above, so I will address his concerns, point by point.
- H3K27 methylation and demethylation are global effects. H3K27 methylation may potentially turn off or down-regulate thousands or tens of thousands of genes. And demethylation of H3K27me loci via JMJD2 can potentially upregulate the expression of the same thousands or tens of thousands of genes.
the key to seeing this is understanding the profound difference between gene promoter site methylation and histone methylation. Both are in the DNA. Individual gene promoter site (or individual gene promoter site neighborhood) methylation is what is involved in the aging clocks promoted by Steve Horvath and others. In general, when a gene is methylated its expression is inhibited. That is, transcriptional repression is associated with DNA methylation which occurs by the covalent modification of cytosine residues in CpG dinucleotides concentrated in CpG islands.
Histone methylation is a completely different matter involving very many genes. H3K27 is the address of a particular histone tail on a nucleosome, namely lysine27 on histone 3. It is not the name or address of any particular gene. When that position is methylated, or in the case of H3K27me doubly or trippily methylated, the chromatin in that neighborhood becomes closed, meaning that genes in that neighborhood cannot be expressed. How many genes are therefore affected? Going back to basics, “A single nucleosome consists of about 150 base pairs of DNA sequence wrapped around a core of histone proteins. The nucleosomes are arranged like beads on a string. They are repeatedly folded in on themselves to form a chromosome (ref).”
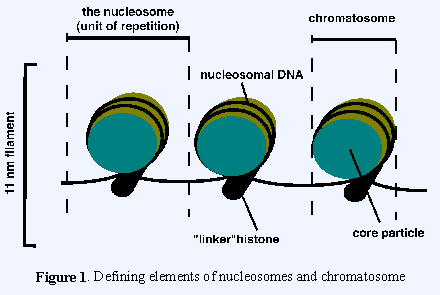
There are 21 human chrosomes and each one may have hundreds of thousands of nucleosomes(ref), and a single human cell might have 30 million nucleosome’s(ref). So it is simple arithmetic to see that this one simple kind of histone methylation, H3K27, could potentially silence thousands or tens of thousands of our genes. Likewise, demethylating H3K27 via JMJD2/ JMJD3 could reactivate the same genes. So yes, one single demethylase could have a massive global effect.
- What does methylation and demethylation of H3K27me histones do?
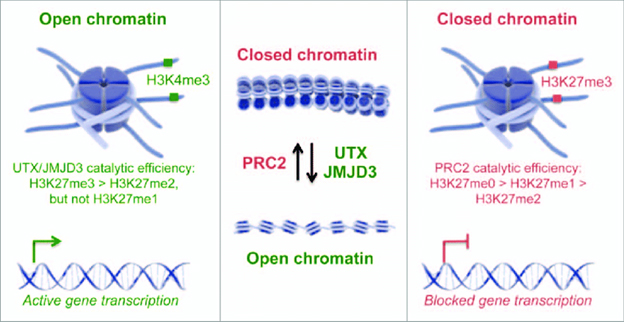
In general: all Methylation and demethylation of histones turns the genes in neighboring DNA “off” and “on,” respectively, either by wrapping the DNA histone tails tightly around the DNA and thereby restricting access to it and its restricting its activation, or by loosening their tails, thereby allowing transcription factors and other proteins to access the DNA. Put differently histone methylation results in closed chromatin where genes are not expressed and demethylation results in open chromatin where genes are expressed.
Histone methylation is one of the several forms of post-translational modifications of chromatin. A reminder of background on such modifications. If the DNA in a typical human cell were stretched out end to end, it would be roughly 6 feet long. “This remarkable feat is accomplished through the intricate organization of that DNA into chromatin. The organization and structure of chromatin can control gene expression. — The fundamental unit of chromatin consists of DNA wrapped around protein octamers, termed histones, in 147 base pair segments to form nucleosome subunits. Histone octamers are made of two copies of each core histone: H2A, H2B, H3, and H4. These histones have positively charged amino (N)-terminal tails which extend from the nucleosome and can undergo several modifications, which in turn affect chromatin accessibility and gene expression.” These modifications, include acetylation, methylation, and ubiquitination, among others, and are regulated by various chromatin-modifying enzymes, frequently referred to as “writers” and “erasers,” which are responsible for incorporating or removing modifications, respectively. In addition to histones, writer and eraser proteins can also interact with transcription factors and other proteins, allowing for an incredibly intricate and multilayered system for the fine-tuned regulation of gene expression(ref).”
Going further: “This structure allows large amounts of DNA to be packaged into a relatively small nucleus. The nucleosome is highly modifiable, which allows the DNA to be accessible or inaccessible for transcription [4]. Post-translational modifications of the chromatin determine accessibility. — The histones are globular in nature except for their tails. It is in the histone tails that there can be multiple modifications of which at least eight are now known. These histone tail modifications include methylation, acetylation, phosphorylation [5], ubiquitination, citrullination, sumoylation and adenosine diphosphate ribosylation [6]. Furthermore, there are families of enzymes that mediate these histone modifications, such at the histone acetyltransferases (HATs) that catalyse the addition of an acetyl group from a donor, acetyl-CoA. Hyperacetylation of histone tails results in opening up of the DNA and thus permits access to transcription factors promoting gene expression. There are also enzymes that remove the modifications from the histone tails, such as the histone deactylases (HDACs), which remove the acetyl groups leading to a closed chromatin structure and therefore gene repression [7]. Acetylation is by far the most common histone modification.(reference: Epigenetic modulation as a therapy in systemic sclerosis).” See also Readers, Writers, and Erasers – Chromatin as the Whiteboard of Heart Disease.
In this blog entry we are particularly concerned with histone methylation. In general, I will continue to cite facts and publications here which indicate that JMJD2/3 and other members of the Jumonji complex result in the activation of numerous needed growth and development and health-related genes, but like so many other substances in biology, which promote growth and development are also correlated with cancer development.
Clarification of these matters can be found in the 2014 publication The Jumonji family: past, present and future of histone demethylases in cancer.
- JMJD2 is a highly specific histone deacetylase that localizes to H3K27me histone sites. It demethylates the gene promoters near such sites.
B. Histone methylation, particularly that related to H3K27me has for some time been known to be related to longevity. The 2011 review Histone methylation makes its mark on longevity relates: “How long organisms live is not entirely written in their genes. Recent findings reveal that epigenetic factors that regulate histone methylation, a type of chromatin modification, can affect lifespan. The reversible nature of chromatin modifications suggests that therapeutic targeting of chromatin regulators could be used to extend lifespan and healthspan. This review describes the epigenetic regulation of lifespan in diverse model organisms, focusing on the role and mode of action of chromatin regulators that affect two epigenetic marks, trimethylated lysine 4 of histone H3 (H3K4me3) and trimethylated lysine 27 of histone H3 (H3K27me3), in longevity.
C. JMJD2 is essential all for the renewal of blood cells. This is explained in the publication The KDM4/JMJD2 histone demethylases are required for hematopoietic stem cell maintenance. “Taken together, our results show that the KDM4 demethylases are required for the expression of genes essential for the long-term maintenance of normal hematopoiesis.” {Recall that KDM4 is another name for JDJM2. in fact, there are multiple proteins in both the KDM and JDJM families (ref)}.
D. Continuing expression of JMJD2 is essential for the development of organisms and cell and organ renewal. It is also needed for stem cell pluripotency. This fact is central for the current blog discussion and is highlighted in the 2016 research publication Continual removal of H3K9 promoter methylation by Jmjd2 demethylases is vital for ESC self-renewal and early development. “Chromatin-associated proteins are essential for the specification and maintenance of cell identity. They exert these functions through modulating and maintaining transcriptional patterns. To elucidate the functions of the Jmjd2 family of H3K9/H3K36 histone demethylases, we generated conditional Jmjd2a/Kdm4a, Jmjd2b/Kdm4b and Jmjd2c/Kdm4c/Gasc1 single, double and triple knockout mouse embryonic stem cells (ESCs). We report that while individual Jmjd2 family members are dispensable for ESC maintenance and embryogenesis, combined deficiency for specifically Jmjd2a and Jmjd2c leads to early embryonic lethality and impaired ESC self-renewal, with spontaneous differentiation towards primitive endoderm under permissive culture conditions. We further show that Jmjd2a and Jmjd2c both localize to H3K4me3-positive promoters, where they have widespread and redundant roles in preventing accumulation of H3K9me3 and H3K36me3. Jmjd2 catalytic activity is required for ESC maintenance, and increased H3K9me3 levels in knockout ESCs compromise the expression of several Jmjd2a/c targets, including genes that are important for ESC self-renewal. Thus, continual removal of H3K9 promoter methylation by Jmjd2 demethylases represents a novel mechanism ensuring transcriptional competence and stability of the pluripotent cell identity.”
E. As usual, the situation is complex. JMJD2 is one member of the Jumonji family (Jmjc) demethylases. Some relevant publications cited here relate to JMJD3 instead of or in addition JMJD2. “The JmjC family comprises 30 members that share a JmjC domain. To date, 18 of these have been shown to possess demethylase activity towards H3K4, H3K9, H3K27, H3K36 and H4K20 (8, 17–31). Based on their homology and the presence of different domains, the 30 members can be further classified into sub-families that often share substrate specificity. — As well as sharing a JmjC domain, all members react with α-ketoglutarate (α-KG) in an Fe(II) ion-dependent manner (Figure 2). However, not all have been shown to be catalytically active. In addition, other domains may also mediate demethylase activity independently of the JmjC domain(ref).“
- We generally know quite a bit about the kinds of genes affected by H3K27 methylation and correspondingly by H3K27 demethylation as a result of JMJD2 or JMJD3 expression.
For one thing, this methylation has much to do with the growth and development. l “Methylation of lysine 27 on histone 3 (H3K27me), a modification usually associated with gene repression, has established roles in regulating the expression of genes involved in lineage commitment and differentiation.(ref)” Deregulation of this methylation also appears to play a major role in cancer.
1.4. Inflammation appears to play a major role in creating some forms of global methylation, particularly ones that can lead to forms of cancer.
See Methylation of Polycomb target genes in intestinal cancer is mediated all by inflammation and H3K27 Methylation: A Focal Point of Epigenetic Deregulation and The Jumonji family: past, present and future of histone demethylases in cancer. This is why we conjecture that suppression of systemic inflammation may be an aspect of averting global methylation. (Frequent readers of this blog know of my many published articles relating to the control of chronic inflammation as a strategy for antiaging, and my creation and promotion of a dietary substance focused on that objective. Check out my publications in the last two years.)
- We know of numerous specific substances that activate the expression of JMJD2. Many of these all are quite familiar and long-known to be health and/or longevity- producing.
I only discuss only one familiar example substance here – vitamin C. However, there are several additional old-friend substances including curcumin and high AKBA Boswellia. I expect to discuss use of these and other practical approaches to JMJD2 activation as an approach to YOUNGING in a future blog entry.
-
- Vitamin C promotes the expression of See the 2017 publication Vitamin C enhances the expression of IL17 in a Jmjd2–dependent manner. “Previously, we reported that vitamin C facilitates the CpG demethylation of Foxp3 enhancer in CD4+Foxp3+ regulatory T cells (Tregs) by enhancing the activity of a DNA demethylase ten-eleven-translocation (Tet). However, it is not clear whether vitamin C affects other helper T cell lineages like T helper type 17 (Th17) cells which are related with Tregs. Here, we show that the expression of interleukin-17A (IL17) increases with the treatment of vitamin C but not with other antioxidants. Interestingly, the upregulation of IL17 was not accompanied by DNA demethylation in Il17 promoter and was independent of Tet enzymes. Rather, vitamin C reduced the trimethylation of histone H3 lysine 9 (H3K9me3) in the regulatory elements of the Il17 locus, and the effects of vitamin C were abrogated by knockdown of jumonji-C domain-containing protein 2 (jmjd2). These results suggest that vitamin C can affect the expression of IL17 by modulating the histone demethylase activity.” Note that this is not the only reported association of vitamin C with a YOUNGING process. Vitamin C also enhances epigenetic regression of normal cells towards stem cell status via the OSKM factors(ref).
- “Vitamin C, by enhancing the catalytic activity of JHDMs and TETs drive histone and DNA demethylation in somatic cells that allow pluripotency genes to turn on while simultaneously erasing the epigenetic memory of the adult cell state. — Vitamin C plays a pivotal role in remodeling the epigenome by enhancing the activity of Jumonji-C domain-containing histone demethylases (JHDMs) and the ten-eleven translocation (TET) proteins. By maintaining differentiation plasticity in culture, vitamin C also improves the quality of tissue specific stem cells derived from iPSCs that are highly sought after for use in regenerative medicine. The ability of vitamin C to potentiate the activity of histone and DNA demethylating enzymes also has clinical application in the treatment of cancer. Vitamin C deficiency has been widely reported in cancer patients and has recently been shown to accelerate cancer progression in disease models. Therapies involving high-dose vitamin C administration are currently gaining traction in the treatment of epigenetic dysregulation, by targeting aberrant histone and DNA methylation patterns associated with cancer progression (ref Reprogramming the Epigenome With Vitamin C).”
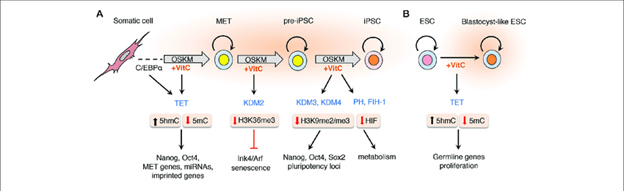
Image source “Vitamin C promotes somatic cell reprogramming by enhancing the activity of α-KGDDs. The addition of vitamin C to the culture medium of somatic cells during reprogramming enhances the activity of α-ketoglutarate dependent dioxygenases (α-KGDDs) including Jumonji-C domain-containing histone demethylases (JHDMs/KDMs), ten-eleven translocation (TET) proteins, prolyl hydroxylases (PH) and the asparaginyl hydroxylase FIH-1. —-loci. The hypomethylation of histones by JHDMs such as KDM2 targets H3K36me3 for demethylation that suppresses the expression of senescence-inducing factors Ink4/Arf. Vitamin C also increases loss of H3K9me2/me3 by enhancing KDM3/4 activity to maintain expression at pluripotency loci during the final stages of reprogramming pre-iPSCs in to fully pluripotent iPSCs.”
5. JMJD3 plays surprising metabolic role, acting together with SIRT1 (itself a gene deacetylase) and PPARα in certain cells under certain circumstances.
The 2018 publication Fasting-induced JMJD3 histone demethylase epigenetically activates mitochondrial fatty acid β-oxidation reports: “Jumonji D3 (JMJD3) histone demethylase epigenetically regulates development and differentiation, immunity, and tumorigenesis by demethylating a gene repression histone mark, H3K27-me3, but a role for JMJD3 in metabolic regulation has not been described. SIRT1 deacetylase maintains energy balance during fasting by directly activating both hepatic gluconeogenic and mitochondrial fatty acid β-oxidation genes, but the underlying epigenetic and gene-specific mechanisms remain unclear. In this study, JMJD3 was identified unexpectedly as a gene-specific transcriptional partner of SIRT1 and epigenetically activated mitochondrial β-oxidation, but not gluconeogenic, genes during fasting. Mechanistically, JMJD3, together with SIRT1 and the nuclear receptor PPARα, formed a positive autoregulatory loop upon fasting-activated PKA signaling and epigenetically activated β-oxidation–promoting genes, including Fgf21, Cpt1a, and Mcad. Liver-specific downregulation of JMJD3 resulted in intrinsic defects in β-oxidation, which contributed to hepatosteatosis as well as glucose and insulin intolerance. Remarkably, the lipid-lowering effects by JMJD3 or SIRT1 in diet-induced obese mice were mutually interdependent.”
- The expression of JMJD2 is not always beneficial. It is highly expressed in many cancers and a cancer therapy research focus is on how to suppress the depression of JMJD2.
See, for example, the 2013 review publication KDM4/JMJD2 Histone Demethylases: Epigenetic Regulators in Cancer Cells and The oncogenic potential of Jumonji D2 (JMJD2/KDM4) histone demethylase overexpression. Also, the 2020 publication Advances in histone demethylase KDM4 as cancer therapeutic targets.
The 2019 publication Histone Modifications as an Intersection Between Diet and Longevity reports: ”Histone modifications are key epigenetic regulators that control chromatin structure and gene transcription, thereby impacting on various important cellular phenotypes. Over the past decade, a growing number of studies have indicated that changes in various histone modifications have a significant influence on the aging process. Furthermore, it has been revealed that the abundance and localization of histone modifications are responsive to various environmental stimuli, such as diet, which can also affect gene expression and lifespan. This supports the notion that histone modifications can serve as a main cellular platform for signal integration. Hence, in this review we focus on the role of histone modifications during aging, report the data indicating that diet affects histone modification levels and explore the idea that histone modifications may function as an intersection through which diet regulates lifespan. A greater understanding of the epigenetic mechanisms that link environmental signals to longevity may provide new strategies for therapeutic intervention in age-related diseases and for promoting healthy aging.”
I expect to include a more systematic discussion of approaches to inducing YOUNGING1.0 in a follow-up blog entry. In particular, I intend to explore the role of histone methylation in mediating the impacts of dietary interventions on aging and age reversal. And I will suggest practical dietary interventions which I believe can contribute to YOUNGING1.0 age reversal.


Thank you for this great article.
I would like to ask you about that here: https://aviv-clinics.com/
Totally different angle, what do you think about it? I am looking for your advice :).
Pingback: YGD YOUNGING1.0 PART 4: UNDERLYING MECHANISMS OF YOUNGING 1.0. HSC STEM CELL DIFFERENTIATION - AGINGSCIENCES™ - Anti-Aging Firewalls™AGINGSCIENCES™ – Anti-Aging Firewalls™
Pingback: YGB YOUNGING1.0 – THE EMERGING AGING REVERSAL STRATEGY - AGINGSCIENCES™ - Anti-Aging Firewalls™AGINGSCIENCES™ – Anti-Aging Firewalls™
Pingback: YGA Introduction to the YOUNGING Series – Emerging Aging Reversal Strategies and Treatments - AGINGSCIENCES™ - Anti-Aging Firewalls™AGINGSCIENCES™ – Anti-Aging Firewalls™