By James P Watson with editorial assistance and a few comments by Vince Giuliano
This is Part 2 of 3 in the series The View from the Telomere end of the Chromsome. Because of the length of the extended telomere story, we subdivided the discussion into three parts. Part 1 relates to a number of more-practical and less-technical topics: the history of telomere biology, telomere length testing, telomerase Inhibitors for cancer, and supplements that activate telomerase and their possible roles for inhibiting or preventing cancers. This Part 2 deals in finer detail with new discoveries related to the molecular biology of telomeres, Sections 1-9. Part 3 relates to additional newer discoveries, Sections 10-17. It also includes a concluding discussion of some of the implications for a Grand Unified Theory of biology of Aging and Biology, as viewed from the ends of a chromosome.
Rather than focusing on telomerase inhibition for cancer and telomerase activation for longevity, scientists are trying to unravel all of the mysteries of why telomere length varies greatly among species, why variations between humans occurs, the relationship of telomeric activities and miotochondrial processes, and the roles of telomeres with respect to the DNA damage response (DDR).
Here I will go over some fascinating new discoveries about telomeres, such as the Shelterin protein Rap1, more about the telomeric long non-coding RNA called TERRA, how telomeric DNA is silenced without DNA methylation, and how sub-telomeric DNA silencing occurs with telomere shortening (aka the Telomere Position Effect, TPE). I will also go over the role of SIRT6 in telomere silencing as well as the non-telomeric roles of SIRT6. I will also go over the newly discovered role of HDAC5 in long telomeres (HDAC 5 is not a Sirtuin protein). Then I will go over the newly discovered roles of the “big four metabolites” in the regulation of chromatin remodeling and DNA repair (Acetyl-CoA, NAD, SAM, FAD, α-KG). I will in the process discuss signaling relating telomeres and mitochondria, and how telomeres are involved in the DNA damage response. I will, in Part 3, discuss some of the large-scale structures that have been recently found in the nucleus called “Epigenetic Regulator Elements” that counteract the TPE (Insulators, MARs, and UCOEs). These epigenetic regulator elements can affect gene expression in new ways that were previously unknown. Ultimately, all of these 17 new discoveries help to explain why we age. I hope the readers of Aging Sciences will enjoy this new look at telomere biology and appreciate all of the complexities that have been discovered. To bookend this discussion, I will both introduce this Part 2 blog entry and end Part 3 with comments relating telomere biology to an emerging Grand Unified Theory of Aging and Biology (GUTab).
Telomere Biology as Part of a Grand Unified Theory of Aging and Biology(GUTab)
The readers of this blog will know that Vince Giuliano is a strong proponent of creating a unified, integrated theory of biology that includes all of the many aspects of aging seen in the laboratory and in nature. Dr. Giuliano has referred to this concept as a “Grand Unified Theory of Aging and Biology” (GUTab). I completely agree with this view and believe that telomere biology must be included in this GUTab as part of a cohesive theory. Grand Unified Theories must unite and simplify things, not create more complex explanations. (For instance, a very elegant theory in physics is the simple equation, E = mc2). In this respect, this blog has so far fallen short of this goal and failed to simplify telomere biology. Nevertheless, I have here laid out some 16 major recent discoveries as a list and will conclude the Part 3 entry with an initial attempt to unify all 16 concepts into a GUTab from the viewpoint of telomeres and aging. Vince and I both believe that much like the universe, “there is no center to the molecular biology of aging”. At the same time, we believe that viewing aging from many angles will help us understand “the beast”, much like the different descriptions of the elephant by the 6 blind men in John Godfrey Saxe’s classical poem The Blind Men and the Elephant.
Now I will go on with the 17 major new discoveries in Telomere and subtelomere biology.
1. Telomeric Loss of Silencing due to lack of SIRT6 activity and how telomeres can be lengthened safely. (i.e. How Dean Ornish lengthened telomeres 10% without TA-65)
Histones silence telomeric and subtelomeric DNA by hypoacetylation and hypermethylation of H3 and H4 histone tails. Heterchromatin protein 1 (HP1) also bind to H3K9me3 at both telomeres and subtelomeric DNA, re-enforcing the “silencing signature”. (HP1α, HP1β shown below on the H3K9 trimethylated lysine residues). There are actually two histone silencing signatures at telomeres and subtelomeric DNA:
1) H3 lysine#9 trimethylation (H3K(me3) – this is the one that HP1 likes
2) H4 lysine #20 trimethylation (H4K20me3) shown in diagram below.
As telomeres shorten with cell division, the heterochromatin marks are lost when SIRT6 is not working due to NAD+ deficiency or due to a lack of SIRT1 (SIRT1 binds to the promoter for SIRT6). SIRT6 normally deacetylates subtelomeric or telomeric DNA. When no deacetylation occurs, the sequence illustrated below occurs, which is called “aging step 1”. However, when the open chromatin structure occurs with acetylation, this allows for telomere elongation to occur by either Telomerase enzyme or the ALT pathway (recombination). Once telomeres are sufficiently elongated, they can be assembled into hetrochromatin by SUV39H or SUV4-20H histone methyltransferases, as well as other heterochromatinizing activities, such as Rb proteins, HP1 binding, and DNA methyltransferases (DNMT3b). This is how Dean Ornish and Elizabeth Blackburn were able to get elderly prostate cancer survivors to lengthen their telomeres by 10% over 5 years with exercise, low fat vegan diet, fish oil, tofu, soy protein drink, selenium supplement, Vitamin C, and Vitamin E .
Reference: Effect of comprehensive lifestyle changes on telomerase activity and telomere length in men with biopsy-proven low-risk prostate cancer: 5-year follow-up of a descriptive pilot study (September 2013).
The diagram below illustrates the loss of silencing that occurs with telomere shortening and SIRT6 lack of activity:

Image source The left-hand arrow shows aging cell-division acetylation affects on telomeres and subtelomeric regions. The right hand arrow shows restorative effects enabled by the open chromatin due to acetylation.
2. TERRA – The Long non-coding RNA that directly inhibits telomerase
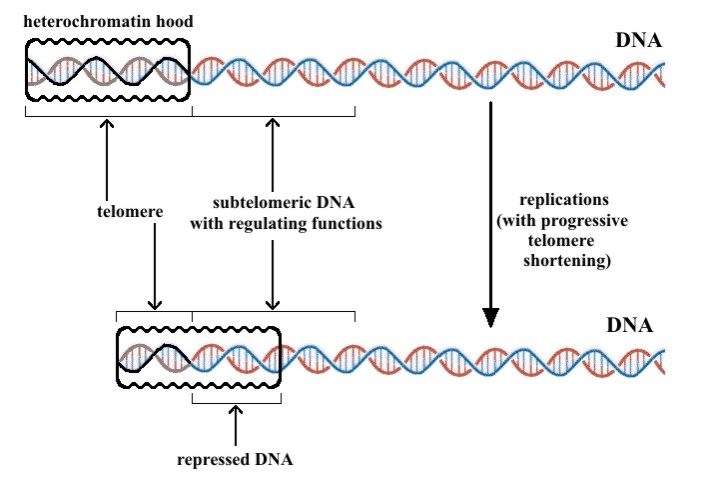
The March 2011 blog entry The epigenetic regulation of telomeres offers an introduction to telomere biology and how they are regulated and to the important role of TERRA. However, much important insight related to telomeres has been discovered in just the last 3 years. For example, there is increasing evidence that subtelomeric DNA is silenced with aging and that this may be an early event that occurs in cells, long before cell cycle arrest or apoptosis occurs. This subtelomeric DNA and how it is silenced is a hot topic right now in aging. This illustration shows where the subtelomeric DNA is:

Not all telomeric DNA is silenced by H3K9me3 + HP1 +H4K20me3 heterochromatin marks.
Some of the telomeric repeats (TTAGGG) are transcribed into a long non-coding RNA, called TERRA, from the open, acetylated sections of telomeric DNA. This gives an RNA sequence of UUAGGG repeats. The 3′ end of this long noncoding RNA is complimentary to the template sequence of telomerase RNA (hTR). As a result, TERRA functions as a direct inhibitor of Telomerase (I don’t think TA-65 works to reverse this!). In addition, this TERRA contacts the telomerase reverse transcriptase (TERT) protein subunit independently of hTR. As a result, this long non-coding RNA functions as a competitive inhibitor for telomeric DNA in addition to exerting an uncompetitive mode of inhibition. It appears that two histone marks are associated with TERRA transcription:
1) H3BK5methylation – maybe a mark of active transcription of TERRA
2) H3K4trimethylation – maybe a mark of active transcription of TERRA
Here are some pictures:

Article and picture reference: The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase
“Proposed modes for telomerase sequestration by TERRA. TERRA (red line) base pairs with the telomerase RNA template (U-shaped blue line) and it interacts with the TERT polypeptide (dark-blue rounded rectangle). Three scenarios are modeled. (1) TERRA may be released from the telomere and bind and inhibit telomere-proximal telomerase molecules. (2) Telomere-bound TERRA may bind and sequester telomerase and prevent its access to the telomeric 3′ end. It is unknown how TERRA is bound to telomeric chromatin; telomeric chromatin binding of TERRA is modeled with the gray oval. (3) TERRA may bind to telomeric chromatin bound telomerase and prevents it from accessing the telomeric 3′ end. It is unknown how telomerase is bound to telomeric chromatin; telomeric chromatin binding of telomerase is modeled with the green oval. Human telomerase may be a dimer (see text), but for simplicity it is modeled here as a monomer.”

Article reference for picture “TERRA/TelRNAs associate to telomeric chromatin and may be involved in regulation of telomere length. Model for a role of telomeric RNAs in the regulation of telomere length. TERRA/TelRNA acts as a potent inhibitor of telomerase activity in vitro, possibly by formation of RNA:RNA hybrids with the template region of the telomerase RNA component.”
3. Rap1 – The Cap Protein that Controls Telomere function, Subtelomere gene expression, and genome-wide gene expression
The Shelterin complex is the protein “chromosome cap” that sits on top of the end of the telomere. There are many proteins in the Shelterin complex, but the most important one appears to be Rap1. When the Rap1 gene is deleted in mice, they have shorter telomeres and develop skin hyperpigmentation in adulthood. Recently, Rap1 has been shown to bind to telomeric DNA as well as extratelomeric sites throughout the genome. Specifically, subtelomeric DNA has many binding sites for Rap1. Of the extratelomeric sites for Rap1 binding, 70% are in the vicinity of genes. When the Rap1 gene was deleted, 31% of the genes with Rap1-binding sites were deregulated. Rap1 protects chromosome ends by blocking the non-homologous end joining (NHEJ) pathway, illustrated below.

Article and illustration reference: The protein Rap1 protects genome integrity by preventing chromosomal fusions “

Reference website for illustration and legend “Telomerase synthesizes the TG-rich simple repeat sequence that provides the DNA platform for its own recruitment and for assembly of the telomere cap. In budding yeast, the Rap1 protein the duplex part, of the telomeric TG repeats, and the Cdc13 protein binds to a 3’ TG-rich single-stranded extension that is a common feature of all telomeres. These two essential proteins organize the telomere cap and regulate the action of telomerase at telomeres so that the TG-repeat tracts are kept within a narrow range of lengths. We proposed the existence of a negative feedback system (often referred to as the “counting model”) in which the number of telomere-bound Rap1 molecules, together with the Rap1-interacting Rif proteins, generates a tract-length dependent signal controlling telomerase.”
4. Telomeric/subtelomeric silencing occurs via a sequence that starts with H3K9 trimethylation, followed by HP1 protein binding to H3K9me3, followed by H4K20 trimethylation, followed by DNMT methylation of DNA CpG cytosines.
The H3K9me3, HP1, and H4K20me3 changes have already been described in Section 1 above. Here we show that these events occur in sequence by the following enzymes:
a) Histone H3K9 trimethylation – this occurs due to Suv39H
b) protein HP1 binding – the H3K9me3 modification creates a binding site for HP1
c) Histone H4K20 trimethylation – this occurs due to Suv4-20H
d) DNA CpG cytosine methylation – this occurs due to DNMTs
As a result of this process, normal subtelomeric DNA is heavily methylated at CpG dinucleotides. Telomeric DNA is NOT methylated, however, since there are no cytosines are found in telomeres (only T,A, and G are found in the telomeric repeats). The methylation of subtelomeric DNA must be very important, however, because when DNA methyltransferases are inactivated in stem cells, the cells loose their telomere length a rapid rates. In cancer, Suv4-20H deficiency occurs sometimes. As a result, there is telomere elongation and derepression of telomere recombination. Here are some diagrams that illustrate this:
article references:
- Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination
- Epigenetic regulatory elements associate with specific histone modifications to prevent silencing of telomeric genes

Picture and legend reference “Model for chromatin assembly at mammalian telomeres and subtelomeres. Both telomeric and subtelomeric chromatin is enriched in trimethylated H3K9, which is performed by the Suv39h HMTases (Garcia-Cao, M., R. O�Sullivan, A.H. Peters, T. Jenuwein, and M.A. Blasco. 2004. Nat. Genet. 36:94-99). In turn, this modification creates an affinity site for the HP1 family of proteins that are important for heterochromatin assembly. In addition, subtelomeric DNA domains are enriched in methylated CpG residues, which are maintained by the Dnmt1, Dnmt3a, and Dnmt3b DNA methyltransferases (Gonzalo, S., I. Jaco, M.F. Fraga, T. Chen, E. Li, M. Esteller, and M.A. Blasco. 2006. Nat. Cell Biol. 8:416-424). Both telomeres and subtelomeres are also enriched in trimethylated H4K20, which is dependent on the activity of the Suv4-20h HMTases (this study). The Suv4-20h enzymes are recruited to chromatin through their interaction with the HP1 proteins (Schotta, G., M. Lachner, K. Sarma, A. Ebert, R. Sengupta, G. Reuter, D. Reinberg, and T. Jenuwein. 2004. Genes Dev. 18:1251-1262). Abrogation of Suv39h or Suv4-20h HMTases lead to the loss of telomere length control, increased telomere recombination, and increased APB frequencies independently of DNA methylation (this study). In turn, loss of DNA methylation can lead to the loss of telomere length control, increased telomere recombination, and increased APB frequencies independently of histone methylation (Gonzalo, S., I. Jaco, M.F. Fraga, T. Chen, E. Li, M. Esteller, and M.A. Blasco. 2006. Nat. Cell Biol.8:416-424).”
5. The “Telomere Position Effect” in Yeast
In yeast, telomeres are silenced by three Sirtuin proteins: Sir2, Sir3, and Sir4. This complex of three Sirtuins silences telomeric DNA and subtelomeric DNA, as illustrated in A below. However, when Sir3 levels are high, Sir3 spreading occurs and silences more subtelomeric DNA. As a result, active chromatin can be silenced, which is called the “Telomere Position Effect” or TPE, illustrated in B below. In yeast, Sir3 also results in “clustering” of the silencing signature (I did not include this in the picture below, but is in the article). This accounts for the silencing of genes near telomeres in yeast, but is not the mechanism of subtelomere gene silencing in humans. In Part 3 of this series, we will discuss three compensatory mechanisms that prevent the TPE.
Article refence:
Clustering heterochromatin: Sir3 promotes telomere clustering independently of silencing in yeast

6. The “Telomere Position Effect” in Mammalian Cells – Repairing “Split Ends” (aka The Dual Role of SIRT6 in repairing DNA and silencing telomeres)
In mammalian cells, there is no Sir2/Sir3/Sir4 complex on telomeric and subtelomeric DNA. Instead, the complexes are as was described in sections 1 and 3 above. Sirtuins do have a role in telomere biology in humans, however. SIRT6 has two roles based on its two functions – deacetylation of histones and mono-ADP ribosylation of PARP1 proteins on Histone 3 subunits. SIRT6 deacetylates the H3K9 and H3K56 lysines. This increases telomere stability and also promotes double stranded DNA break repair by both the HR and NHEJ routes via PARP1 independent routes. (Illustrated below) In addition, SIRT6 puts multiple mono-ADP-ribosyl groups on the DNA repair protein, PARP1, which also repairs DNA in response to oxidative stress. Unfortunately, all of this DNA repair by PARP1 uses up NAD+, which leaves the cell nuclear levels of NAD+ low. As a result, all of the SIRT1 proteins cannot work since they all need NAD+ as a cofactor for enzyme action. Thus, the dual role of SIRT6 in both telomeric silencing and DNA repair is believed to be a root issue associated with aging.
reference for article with image below: Repairing split ends: SIRT6, mono-ADP ribosylation and DNA repair

“SIRT6 regulates genomic stability. SIRT6 promotes genome stability by regulating DNA single-strand and double strand break repair pathways and by facilitating telomere maintenance. The deacetylase and the mono-ADP ribosyltransferase activities of SIRT6 both contribute to this function.” A key issue appears to be whether there is enough of it to satisfy the dual roles.
7. How the TPE could regulate Subtelomeric DNA
Normally in the embryo and in the young person, as a cell ends up with a critically short telomere, the cell undergoes apoptosis. However, as one ages, the possibility of the cell undergoing cellular senescence rather than apoptosis (with telomere shortening) increases. No simple explanation for this increased tendency towards cellular senescence vs apoptosis has been clearly established. For this reason, several theories have been proposed by several authors, including Fossel (1996), Villeponteau (1997), Wright, et al (2001) and more recently Libertini (2011). Here they propose that with aging, a gradual silencing of subtelomeric genes occurs due to the TPE (as well as other reasons). Specifically, as the telomere shortens, the Shelterin protein cap slides further down the chromosome or the hood shortens. In either case, the subtelomeric DNA gets silenced and the probability of cellular senescence increases. There is some direct evidence for this theory.
Reference: Phylogeny of Age-Related Fitness Decline in the Wild and of Related Phenomena

“ Telomere oscillates between capped and uncapped conditions. The probability of uncapped condition increases at each duplication in relation to telomere shortening. Uncapped telomere acts as a broken end that can cause an end-to-end joining and a block of cell duplications.”
“The expression of many genes is impaired in relation with telomere progressive shortening. As likely hypothesis, a subtelomeric DNA tract regulates overall cell functionality and its action is impaired by the progressive sliding of the heterochromatin ‘hood’ caused by telomere shortening [Fossel, 2004]”
8. Other roles of SIRT6 in the cell – DS Break Repair, Base Excision Repair, NF-kB gene inhibition, HIF-1a gene inhibition, and inhibition of transcription factors
There are several other roles of SIRT6 in the cell, as illustrated below:
6A. Recruitment of SNF2H to site of DNA damage – This is a PARP1 independent, important function of SIRT6. As you can see in the illustration below, SIRT6 binds to the site of a double-stranded DNA break. THIS OCCURS WITHIN 5 SECONDS OF A DS DNA BREAK. Once SIRT6 arrives, it recruits a a chromatin remodeler called SNF2H and deacetylates Histone 3 at lysine 56 (H3K56) Then the SIRT6/SNF2H complex can recruit p53, BRCA1, and RPA to do the repair work. Thus, SIRT6 is vitally important as a “rapid response element” for fixing DNA double stranded breaks.
Reference for article, image and legend below: SIRT6 Recruits SNF2H to DNA Break Sites, Preventing Genomic Instability through Chromatin Remodeling


“DNA damage is linked to multiple human diseases, such as cancer, neurodegeneration, and aging. Little is known about the role of chromatin accessibility in DNA repair. Here, we find that the deacetylase sirtuin 6 (SIRT6) is one of the earliest factors recruited to double-strand breaks (DSBs). SIRT6 recruits the chromatin remodeler SNF2H to DSBs and focally deacetylates histone H3K56. Lack of SIRT6 and SNF2H impairs chromatin remodeling, increasing sensitivity to genotoxic damage and recruitment of downstream factors such as 53BP1 and breast cancer 1 (BRCA1). Remarkably, SIRT6-deficient mice exhibit lower levels of chromatin-associated SNF2H in specific tissues, a phenotype accompanied by DNA damage. We demonstrate that SIRT6 is critical for recruitment of a chromatin remodeler as an early step in the DNA damage response, indicating that proper unfolding of chromatin plays a rate-limiting role. We present a unique crosstalk between a histone modifier and a chromatin remodeler, regulating a coordinated response to prevent DNA damage.(ref)”
6B. Repair of Single Stranded DNA breaks, inhibition of NF-kB gene transcription, inhibition of HIF-1a gene transcription, and the inhibition of transcription factors


Article and image reference: Chromatin regulation and genome maintenance by mammalian SIRT6
“Chromatin-bound: SIRT6 regulates multiple chromatin-templated processes. A. SIRT6 deacetylates H3K9 and H3K56 at telomeres, and SIRT6 depletion triggers aberrant telomere metabolism. B. SIRT6 enables efficient DSB repair. C. Although the role of SIRT6 in BER has not been clearly defined, the spectrum of DNA damage sensitivities of Sirt6−/− mouse cells and the results of functional complementation assays suggest a potential chromatin-regulatory function for SIRT6 in this repair pathway. D. SIRT6 represses NF-κB and HIF1α target gene expression, and might act in conjunction with other transcription factors (TFs) to regulate additional gene expression programs. Question marks indicate hypothesized or uncharacterized roles for SIRT6(ref).”
6C. Growth Hormone and IGF-1 expression in the brain.
This is a new role for SIRT6 that I did not know about. Also, SIRT6 activation decreases triglyceride synthesis. Thus, SIRT6 deficiency or SIRT6 dysfunction due to nuclear NAD+ deficiency can possibly explain many of features of aging.
Article reference for illustration below: Mammalian Sirtuins and Energy Metabolism
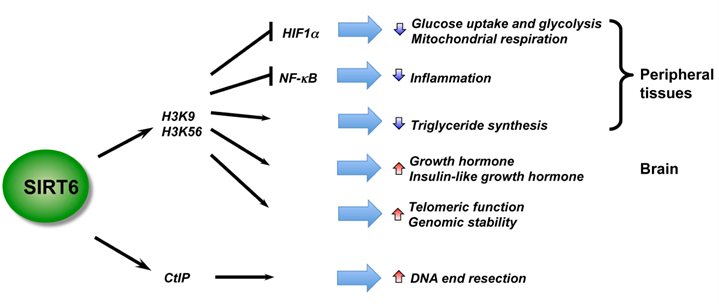

Illustration and legend reference “SIRT6 is a critical regulator in genome stability, metabolism, and inflammatory response. By deacetylation of H3, SIRT6 regulates metabolic homeostasis and inflammatory response in peripheral tissues, while functioning as a central regulator of somatic growth.”
9. Metabolites Control Chromatin Remodeling and DNA repair
Although we typically think that epigenetics (chromatin remodeling) is an “upstream event” and therefore would not be affected by metabolites, the opposite is true. There are at least 5 metabolites whose levels can control various chromatin remodeling pathways and DNA repair mechanisms. This is nicely illustrated below. This diagram shows the main four metabolites:
1. Acetyl-CoA – The Key Acetylator activator
Acetyl-CoA is a required co-factor for histone acetylation. Acetyl-CoA dependent Histone acetylation is a central aspect of the DNA damage response. Citrate is the inhibitor of this reaction. Acetyl-CoA facilitates the relaxation of chromatin around a double stranded DNA break and repair factor access
Reference: http://www.frontiersin.org/journal/10.3389/fgene.2013.00182/full Metabolic modulation of chromatin: implications for DNA repair and genomic integrity
2. NAD+ – The Key Deacetylator activator
NAD+ is a required cofactor for histone deacetylation. NAD+ dependent Histone deacetylation is an important part of DNA repair. NADH is the inhibitor of this reaction. Both SIRT1 and SIRT6 are recruited to sites of DNA damage. SIRT6 deacetylates H3K56ac and SIRT1 deacetylates H3K16ac. Both SIRT6 and SIRT1 can deacetylate H3K9ac.
3. SAM – The Key Methylator Activator
SAM
is a required methyl donor for histone methylation SAM is a key to histone methylation that is required with chromatin remodeling and DNA repair. Methionine is the inhibitor of this reaction. Just like all acetylation reactions are dependent on Acetyl-CoA, all methylation reactions are dependent on SAM. All methyl group additions require the intermediate, SAM H4K20 methylation is one of the key chromatin marks for histone-based gene silencing of subtelomeric DNA and telomeric DNA .This requires SAM
4. FAD and alpha-ketoglutarate – The Key Demethylator Activator
Two co-factors have been found that are necessary for histone demethylation – FAD and a-KG. FAD is the co-factor required for the lysine demethylases 1 and 2 (LSD1, LSD2) alpha-ketoglutarate (a-KG) is the required cofactor for the JmjC domain containing deoxygenases, which is the largest class of histone demethylases. 2-HG is an inhibitor of these reactions.
The diagram below illustrates the four metabolites that control chromatin remodeling and DNA repair. Note that p53 is involved in all four mechanisms. Also note that BRCA1 is involved in two of them: Citrate/Acetyl-CoA and NADH/NAD
Reference for article and image below and legend: Metabolic modulation of chromatin: implications for DNA repair and genomic integrity


“ Cross-talk between metabolites, chromatin and DSB repair. (A) The deposition of chromatin marks is influenced by metabolites, which function as cofactors or substrates for the indicated chromatin modifiers (CMs). Central pathways affecting histone acetylation, deacetylation, PARylation, and histone/DNA methylation and demethylation are shown, arrows indicate positive regulation/deposition of chromatin marks, blunted arrows depict negative regulation/removal of chromatin marks. (B)Chromatin marks deposited or removed through pathways in (A) have been implicated in the modulation of the recruitment of the indicated repair factors, suggesting a possible link between metabolites and DSB repair. Green arrows indicate that a given chromatin modification results in increased recruitment/activation of the indicated repair factors, red arrows depict impaired activation/recruitment in the presence of the same modification. See Table 1 for a detailed list of modifiers and modifications involved in DSB repair. In addition to chromatin, the HAT Tip60 can acetylate and activate the central DDR mediator ATM kinase (gray arrows), whereas ATM activity is negatively affected by DNA methylation (blunted gray arrow). Ac, acetylated histone; me, methylated histone; Cme, methy-cytosine; PAR, poly-(ADP-ribose); ETC, electron transport chain; HAT, histone acetyl transfrease; HMT, histone methyltransferase; KDM, lysine demethylase. See text for enzyme and metabolite abbreviations.
On to Part 3 of this View from the Telomere end of the Chromosome series
Topics that will be discussed in Part 3 include:
10. Regulatory Elements Prevent Silencing of Subtelomeric Genes
11. Insulators – The partitioners of the genome
12. MARs – Attachement Points for DNA to the Nuclear Cytoskeleton
13. UCOEs – A Promoter-based method of stabilizing gene expression
14. HDAC5 and Telomeres – Required for maintaining normal telomeres
15. How Caloric Restriction and p53 can activate Telomere Maintenance Genes and DNA repair genes
16. Relationship of telomere dysfunction and mitochondrial dysfunction
17. Cross-talk between sirtuins
Towards a GUT, looked at from the end of the chromosome


Pingback: A Dietary Supplement That Can Increase Longevity? - The Cancer Expert